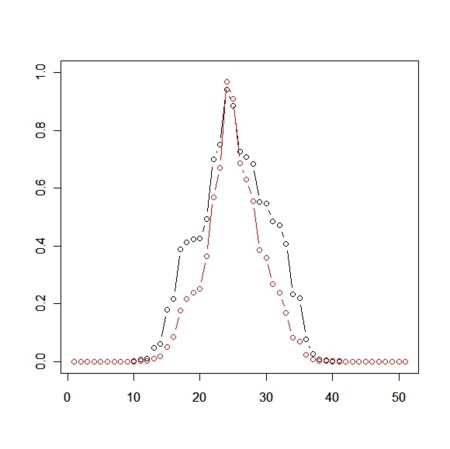
(8) 量的形質の増減(paired t-test)
# paired test N<-50 par(ask=TRUE) Pre<-rnorm(N) d<-runif(N)^2 pairedTps<-rep(0,N+1) rankSumps<-rep(0,N+1) for(i in 0:N){ Post<-Pre+d*c(rep(-1,i),rep(1,N-i)) plot(rep(1,length(Pre)),Pre,pch=18,col=3,xlim=c(0,3),ylim=range(c(Pre+max(d),Pre-max(d))),main="",xlab="",ylab="") par(new=TRUE) plot(rep(2,length(Post)),Post,pch=18,col=4,xlim=c(0,3),ylim=range(c(Pre+max(d),Pre-max(d))),main="",xlab="",ylab="") for(j in 1:N){ segments(1,Pre[j],2,Post[j]) } pairedTps[i+1]<-t.test(Pre,Post,paired=TRUE)$p.value rankSumps[i+1]<-wilcox.test(Pre,Post,paired=TRUE)$p.value } plot(pairedTps,type="b",ylim=c(0,1),main="",xlab="",ylab="") par(new=TRUE) plot(rankSumps,col=2,type="b",ylim=c(0,1),main="",xlab="",ylab="")
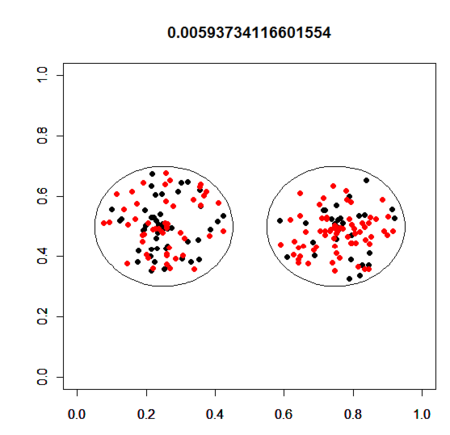
(1) 2x2分割表(ケース・コントロール スタディ)
# 2x2分割表 NOp<-FALSE N1<-100 N2<-100 a<-50 A<-c(rep(1,a),rep(2,N1-a)) xlim<-ylim<-c(0,1) bs<-seq(from=1,to=N2-1,by=1) t<-seq(from=0,to=1,length=100)*2*pi ra<-0.2 rb<-ra max<-0.25 may<-0.5 mbx<-0.75 mby<-0.5 p.out<-rep(0,length(bs)) for(i in 1:length(bs)){ b<-bs[i] B<-c(rep(1,b),rep(2,N2-b)) plot(ra*cos(t)+max,ra*sin(t)+may,type="l",xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(rb*cos(t)+mbx,rb*sin(t)+mby,type="l",xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) rs<-runif(N1)*ra*0.9 ts<-runif(N1)*2*pi plot(rs*cos(ts)+max,rs*sin(ts)+may,pch=19,cex=1,col=A,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) rs<-runif(N2)*rb*0.9 ts<-runif(N2)*2*pi #print(table(as.data.frame(cbind(A,B)))) v1<-c(rep(1,length(A)),rep(2,length(B))) v2<-c(A,B) print(table(v1,v2)) p.f<-fisher.test(table(v1,v2))$p.value p.out[i]<-p.f if(NOp)p.f="" plot(rs*cos(ts)+mbx,rs*sin(ts)+mby,pch=19,cex=1,col=B,xlim=xlim,ylim=ylim,main=p.f,xlab="",ylab="") par(ask=TRUE) } plot(bs,p.out)
第6章 Rでシミュレーション
- 遺伝統計学集中講義のための資料の全体の目次
- Rでシミュレーション
- ここで作ってみるシミュレーション用ソースはジュニアキャンパスのデモ用のものとしてリンクを取る
- ジュニアキャンパスネタは、2012年度の1回生/4回生の講義ネタ
- いろいろな「検定されるべき」データをシミュレーションで作ってみる
- そのうえで「検定する」って何かを考える
(2) 帰無仮説からの2x2表ランダムサンプリング
N1<-100 N2<-150 p1<-p2<-0.3 Niter<-10000 p.out<-rep(0,Niter) v1<-c(rep(1,N1),rep(2,N2)) for(i in 1:Niter){ v2<-sample(c(1,2),N1+N2,replace=TRUE,prob=c(p1,1-p1)) #print(table(v1,v2)) #p.out[i]<-chisq.test(c(rep(1,N1),rep(2,N2)),c(v1,v2))$p.value p.out[i]<-chisq.test(table(v1,v2),correct=FALSE)$p.value } plot(ppoints(Niter,a=0),sort(p.out))
(3) 対立仮説からの2x2表ランダムサンプリング
N1<-100 N2<-150 p1<-0.3 p2<-0.4 Niter<-10000 p.out<-rep(0,Niter) v1<-c(rep(1,N1),rep(2,N2)) for(i in 1:Niter){ v2<-c(sample(c(1,2),N1,replace=TRUE,prob=c(p1,1-p1)),sample(c(1,2),N2,replace=TRUE,prob=c(p2,1-p2))) #print(table(v1,v2)) #p.out[i]<-chisq.test(c(rep(1,N1),rep(2,N2)),c(v1,v2))$p.value p.out[i]<-chisq.test(table(v1,v2),correct=FALSE)$p.value } plot(ppoints(Niter,a=0),sort(p.out))
(4) ケース・コントロール比が大きいとき(コホート疫学)
# コホート # 2x2分割表 NOp<-FALSE N1<-10000 N2<-20 a<-1000 A<-c(rep(1,a),rep(2,N1-a)) A<-sample(A) xlim<-ylim<-c(0,1) bs<-seq(from=1,to=N2-1,by=1) t<-seq(from=0,to=1,length=100)*2*pi ra<-0.2 rb<-ra max<-0.25 may<-0.5 mbx<-0.75 mby<-0.5 p.out<-rep(0,length(bs)) for(i in 1:length(bs)){ b<-bs[i] B<-c(rep(1,b),rep(2,N2-b)) B<-sample(B) plot(ra*cos(t)+max,ra*sin(t)+may,type="l",xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(rb*cos(t)+mbx,rb*sin(t)+mby,type="l",xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) rs<-runif(N1)*ra*0.9 ts<-runif(N1)*2*pi plot(rs*cos(ts)+max,rs*sin(ts)+may,pch=19,cex=0.1,col=A,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) rs<-runif(N2)*rb*0.9 ts<-runif(N2)*2*pi #print(table(as.data.frame(cbind(A,B)))) v1<-c(rep(1,length(A)),rep(2,length(B))) v2<-c(A,B) print(table(v1,v2)) p.f<-fisher.test(table(v1,v2))$p.value p.out[i]<-p.f if(NOp)p.f="" plot(rs*cos(ts)+mbx,rs*sin(ts)+mby,pch=19,cex=1,col=B,xlim=xlim,ylim=ylim,main=p.f,xlab="",ylab="") par(ask=TRUE) } plot(bs,p.out)
(5) 量的形質の分布(t-test)
# 平均を変える N1<-8 N2<-8 m1<-50 m2<-50 v1<-5 v2<-5 d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) mdeltas<-seq(from=-5,to=5,by=0.5) par(ask=TRUE) d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) ylim<-range(c(d1,d2+min(mdeltas),d2+max(mdeltas))) for(i in 1:length(mdeltas)){ #m2<-m2s[i] tmpd2<-d2+mdeltas[i] t.test.out<-t.test(d1,tmpd2) plot(c(rep(1,N1),rep(2,N2)),c(d1,tmpd2),pch=19,cex=0.5,xlim=c(0,3),ylim=ylim,main=t.test.out$p.value,xlab="",ylab="") boxplot(d1,tmpd2,main=t.test.out$p.value,ylim=ylim,xlab="",ylab="") }
(6) 帰無仮説からの量的形質の分布、等分散・異分散(t-test)
- 等分散からのサンプリング
# 帰無仮説 N1<-5 N2<-5 m1<-500 m2<-500 v1<-10 v2<-10 # 異分散 Niter<-1000 ps1<-ps2<-rep(0,Niter) for(i in 1:Niter){ d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) t.test.out1<-t.test(d1,d2,var.equal=TRUE) t.test.out2<-t.test(d1,d2,var.equal=FALSE) ps1[i]<-t.test.out1$p.value ps2[i]<-t.test.out2$p.value } plot(ps1,ps2,cex=0.1) plot(ppoints(Niter,a=0),sort(ps1),cex=0.1) plot(ppoints(Niter,a=0),sort(ps2),cex=0.1)
- 異分散からのサンプリング
# 帰無仮説 N1<-5 N2<-5 m1<-500 m2<-500 v1<-1 v2<-200 # 異分散 Niter<-1000 ps1<-ps2<-rep(0,Niter) for(i in 1:Niter){ d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) t.test.out1<-t.test(d1,d2,var.equal=TRUE) t.test.out2<-t.test(d1,d2,var.equal=FALSE) ps1[i]<-t.test.out1$p.value ps2[i]<-t.test.out2$p.value } plot(ps1,ps2,cex=0.1) plot(ppoints(Niter,a=0),sort(ps1),cex=0.1) plot(ppoints(Niter,a=0),sort(ps2),cex=0.1)
(7) 対立仮説からの量的形質の分布、等分散・異分散(t-test)
- 等分散からのサンプリング
# 対立仮説 N1<-5 N2<-5 m1<-500 m2<-510 v1<-10 v2<-10 # 異分散 Niter<-1000 ps1<-ps2<-rep(0,Niter) for(i in 1:Niter){ d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) t.test.out1<-t.test(d1,d2,var.equal=TRUE) t.test.out2<-t.test(d1,d2,var.equal=FALSE) ps1[i]<-t.test.out1$p.value ps2[i]<-t.test.out2$p.value } plot(ps1,ps2,cex=0.1) plot(ppoints(Niter,a=0),sort(ps1),cex=0.1) plot(ppoints(Niter,a=0),sort(ps2),cex=0.1)
- 異分散からのサンプリング
# 対立仮説 N1<-5 N2<-5 m1<-500 m2<-510 v1<-1 v2<-200 # 異分散 Niter<-1000 ps1<-ps2<-rep(0,Niter) for(i in 1:Niter){ d1<-rnorm(N1,m1,sqrt(v1)) d2<-rnorm(N2,m2,sqrt(v2)) t.test.out1<-t.test(d1,d2,var.equal=TRUE) t.test.out2<-t.test(d1,d2,var.equal=FALSE) ps1[i]<-t.test.out1$p.value ps2[i]<-t.test.out2$p.value } plot(ps1,ps2,cex=0.1) plot(ppoints(Niter,a=0),sort(ps1),cex=0.1) plot(ppoints(Niter,a=0),sort(ps2),cex=0.1)
(9) 2次元の的当て(2次元正規分布)
- 対立仮説が1点
# 2次元の的当て Npt<-50 xlim<-c(-1,1) ylim<-xlim ori<-c(0,0) mx<-0.2 my<-0 sdx<-1/5 sdy<-1/5 r<-1 # 2 は正規分布 rateParam<-0.1 XYs<-matrix(10,Npt,2) for(i in 1:Npt){ plot(ori[1],ori[2],cex=60,pch=19,col=1,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=50,pch=19,col=2,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=40,pch=19,col=3,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=30,pch=19,col=4,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=20,pch=19,col=5,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=10,pch=19,col=6,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) par(new=TRUE) tmpx<-rnorm(1,mean=mx,sd=sdx) tmpy<-rnorm(1,mean=my,sd=sdy) tmpR<-sqrt(tmpx^2+tmpy^2) XYs[i,1]<-tmpx*tmpR^(r/2) XYs[i,2]<-tmpy*tmpR^(r/2) plot(XYs[,1],XYs[,2],cex=1,pch=19,col=gray(100/100),xlim=xlim,ylim=ylim,main="",mex=30,xlab="",ylab="") n0<-i%%10 n1<-((i-n0)/10)%%10 n2<-((i-n0-n1*10)/100)%%10 par(new=TRUE) plot(0.9,0.9,pch=n0+48,cex=4.5,xlim=xlim,ylim=ylim,main="") par(new=TRUE) plot(0.77,0.9,pch=n1+48,cex=4.5,xlim=xlim,ylim=ylim,main="") par(new=TRUE) plot(0.64,0.9,pch=n2+48,cex=4.5,xlim=xlim,ylim=ylim,main="") tmpCurrent<-Sys.time() while(Sys.time()-tmpCurrent<rateParam){ } }
- 中心からある距離の点(ならどこでもよい)を狙っている
# 半径が一定Rtの点を的にする Npt<-500 xlim<-c(-1,1) ylim<-xlim ori<-c(0,0) mx<-0.2 my<-0 sdx<-1/16 sdy<-1/16 Rts<-seq(from=0,to=0.6,by=0.01) for(j in 1:length(Rts)){ Rt<-Rts[j] r<-1 # 2 は正規分布 rateParam<-0.1 XYs<-matrix(10,Npt,2) #for(i in 1:Npt){ i<-Npt plot(ori[1],ori[2],cex=60,pch=19,col=1,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=50,pch=19,col=2,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=40,pch=19,col=3,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=30,pch=19,col=4,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=20,pch=19,col=5,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) plot(ori[1],ori[2],cex=10,pch=19,col=6,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") par(new=TRUE) par(new=TRUE) for(i in 1:Npt){ tmpr<-runif(1)*2*pi mtx2<-Rt*cos(tmpr) mty2<-Rt*sin(tmpr) tmpx<-rnorm(1,mean=mtx2,sd=sdx) tmpy<-rnorm(1,mean=mty2,sd=sdy) tmpR<-sqrt(tmpx^2+tmpy^2) XYs[i,1]<-tmpx*tmpR^(r/2) XYs[i,2]<-tmpy*tmpR^(r/2) } plot(XYs[,1],XYs[,2],cex=1,pch=19,col=gray(100/100),xlim=xlim,ylim=ylim,main="",mex=30,xlab="",ylab="") #n0<-i%%10 #n1<-((i-n0)/10)%%10 #n2<-((i-n0-n1*10)/100)%%10 #par(new=TRUE) #plot(0.9,0.9,pch=n0+48,cex=4.5,xlim=xlim,ylim=ylim,main="") #par(new=TRUE) #plot(0.77,0.9,pch=n1+48,cex=4.5,xlim=xlim,ylim=ylim,main="") #par(new=TRUE) #plot(0.64,0.9,pch=n2+48,cex=4.5,xlim=xlim,ylim=ylim,main="") #tmpCurrent<-Sys.time() #while(Sys.time()-tmpCurrent<rateParam){ #} #} }
(10) ポアソン過程(ランダムに起きること)
- 一定間隔で起きること、まったくでたらめに起きること(どの時点も同じ確率で起きうる〜ポアソン過程、生起間隔が指数分布)、起きるとなったら集中しておきがちなこと
# ぽつりぽつりと起きる現象 m<-0.5 Ne<-20 proportionRunif<-0.3 proportionRnorm<-0.4 events<-rexp(Ne*(1-proportionRunif-proportionRnorm),m) evPows<-c(0.01,0.1,0.5,1,2,3,5) for(ep in evPows){ evPow<-ep events2<-events^evPow cumeve<-cumsum(events2) dur<-20 cumeve<-cumeve/max(cumeve)*dur currentTime<-Sys.time() par(ask=FALSE) for(i in 1:length(cumeve)){ while(Sys.time()-currentTime<cumeve[i]){ } plot(cumeve[1:i],rep(0,i),pch=19,cex=0.5,xlim=c(0,max(cumeve)),main="",xlab="時間",ylab="") } par(ask=TRUE) plot(cumeve[1:i],rep(0,i),pch=19,cex=0.5,xlim=c(0,max(cumeve)),main="",xlab="時間",ylab="") } for(ep in evPows){ evPow<-ep events2<-events^evPow cumeve<-cumsum(events2) dur<-10 cumeve<-cumeve/max(cumeve)*dur currentTime<-Sys.time() xlim<-ylim<-c(-1,1) par(ask=FALSE) for(i in 1:length(cumeve)){ plot(c(10),c(10),pch=19,cex=4,main="",xlab="",ylab="",xlim=xlim,ylim=ylim) while(Sys.time()-currentTime<cumeve[i]){ } plot(c(0),c(0),pch=19,cex=10,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") } par(ask=TRUE) plot(c(0),c(0),pch=19,cex=10,xlim=xlim,ylim=ylim,main="",xlab="",ylab="") }
(11) 有害事象
# 有害事象 N<-500 # 単位期間にDという有害事象が起きる確率 pD<-0.1 D<-runif(N)/pD # 単位期間にVという有害事象が起きる確率 pV<-1 V<-runif(N,min=0,max=1)/pV t<-seq(from=0,to=1,length=100) xlim<-ylim<-c(0,1) plot(rep(0,length(t)),t,xlim=xlim,ylim=ylim,type="l") Thres<-0.1 for(i in 1:N){ #par(new=TRUE) if(D[i]<1){ col<-1 segments(0,i/N,D[i],i/N,col=col) if(D[i]-V[i]<Thres && V[i]<D[i]){ col<-2 segments(V[i],i/N,D[i],i/N,col=col) } points(D[i],i/N,col=1,pch=15) if(V[i]<D[i]){ points(V[i],i/N,col=2,pch=19) } } }
- ある事象が確率的に別の事象を引き起こしがちだというシミュレーション
N<-100 # 単位期間にDという有害事象が起きる確率 pD<-1 D<-runif(N)/pD # 単位期間にVという有害事象が起きる確率 pV<-1 # Vの後、probの確率で、Dが短期間に引き起こされる(有害事象が起きる) V<-runif(N,min=0,max=1)/pV VearlierD<-which(V<D) SideEffectM<-0.01 SideEffectV<-0.00001 prob<-0.5 Sideeffected<-sample(VearlierD,length(VearlierD)*prob) D[Sideeffected]<-V[Sideeffected]+abs(rnorm(length(Sideeffected),SideEffectM,sqrt(SideEffectV))) t<-seq(from=0,to=1,length=100) xlim<-ylim<-c(0,1) for(i in 1:length(t)){ plot(rep(0,length(t)),t,type="l",xlim=xlim,ylim=ylim) for(j in 1:N){ if(t[i]<min(D[j],V[j])){ segments(0,j/N,t[i],j/N) }else{ if(D[j]<V[j]){ segments(0,j/N,D[j],j/N) }else{ segments(0,j/N,V[j],j/N) col<-1 if(t[i]>D[j])col<-2 segments(V[j],j/N,min(t[i],D[j]),j/N,col=col) } } if(D[j]<t[i])points(D[j],j/N,pch=15) if(V[j]<t[i] && V[j]<D[j])points(V[j],j/N,pch=19,col=2) } }
(12) 塩基置換
#(U,C,A,G)=(1,2,3,4) b<-1:4 #b<-c("T","C","A","G") codons<-as.matrix(expand.grid(b,b,b)) aas<-c( "F","F","L","L","S","S","S","S","Y","Y","X","X","C","C","X","W","L","L","L","L","P","P","P","P","H","H","Q","Q","R","R","R","R","I","I","I","M","T","T","T","T","N","N","K","K","S","S","R","R","V","V","V","V","A","A","A","A","D","D","E","E","G","G","G","G" ) nonStops<-which(aas!="X") st<-which(aas=="M") Stops<-which(aas=="X") L<-10 peptides<-c(st,sample(nonStops,L-1,replace=TRUE)) bases<-c() for(i in 1:L){ tmpaa<-peptides[i] candidates<-which(aas==aas[tmpaa]) tmpcodon<-sample(candidates,1) bases<-c(bases,as.vector(codons[tmpcodon,])) } bases base2aa<-function(b,cs){ len<-length(b)/3 ret<-c() for(i in 1:len){ triplet<-b[(i*3):((i-1)*3+1)] for(j in 1:length(cs[,1])){ if(prod(triplet==cs[j,])){ ret<-c(ret,aas[j]) } } } ret } peptides2<-base2aa(bases,codons) Niter<-1000 M<-matrix(0,Niter+1,length(bases)) M[1,]<-bases rt<-0.1 for(i in 1:Niter){ loop<-TRUE while(loop){ rs<-runif(length(bases)) newbs<-bases muts<-which(rs<rt) #print(muts) if(length(muts)>0){ tmpb<-sample(b,length(muts),replace=TRUE) newbs[muts]<-tmpb } #print(bases) #print(newbs) newpeptides<-base2aa(newbs,codons) #print(newpeptides) #print(peptides) if(identical(newpeptides,peptides2)){ loop<-FALSE M[i+1,]<-newbs } } } image(M) M2<-t(t(M)-bases%%4) image(t(sign(M2%%4))) grid(nx=length(M2[1,]),ny=1)