ノンパラメトリック・ベイズ実践編
- Non-parametric bayesian clustering
- Data set simulation
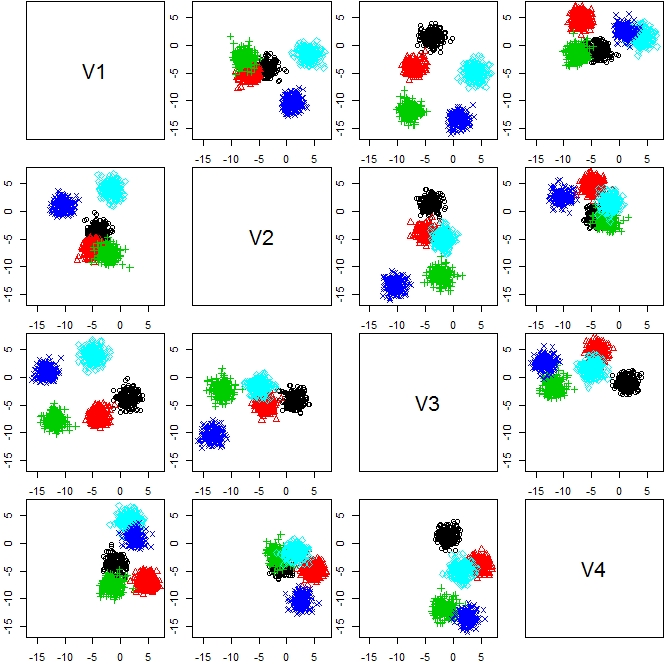
n <- 1000 d <- 4 k <- 5 s <- sample(1:k,n,replace=TRUE) m <- matrix(rnorm(d*k,sd=6),k,d) X <- matrix(0,n,d) for(i in 1:k){ tmp <- which(s == i) r <- matrix(rnorm(d * length(tmp))) X[tmp,] <- r for(j in 1:d){ X[tmp,j] <- X[tmp,j] + m[i,j] } } plot(as.data.frame(X))[f:id:ryamada22:20180922131657j:plain]

library(clues) out <- clues(X) plot(out)
machine-learning.hatenablog.com
を使っているらしく、速い!
# coding: utf-8
# In[1]:
# In[2]:
from sklearn.mixture import BayesianGaussianMixture
from sklearn.datasets import *
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
# In[3]:
plt.style.use("seaborn")
np.random.seed(42)
# In[4]:
X,y = make_blobs(1000,n_features=4, centers = 5)
# In[13]:
dpgmm = BayesianGaussianMixture(n_components=1000)
dpgmm.fit(X)
y_pred = dpgmm.predict(X)
# In[14]:
X_df = pd.DataFrame(X,columns=["var1","var2","var3","var4",])
X_df["label"] = y
X_df["label"] = X_df["label"].astype("str")
X_df["prediction"] = y_pred
X_df["prediction"] = X_df["prediction"].astype("str")
# In[15]:
sns.pairplot(X_df, vars = ["var1","var2","var3","var4",],hue = "label")
plt.show()
# In[16]:
sns.pairplot(X_df, vars = ["var1","var2","var3","var4",],hue = "prediction")
plt.show()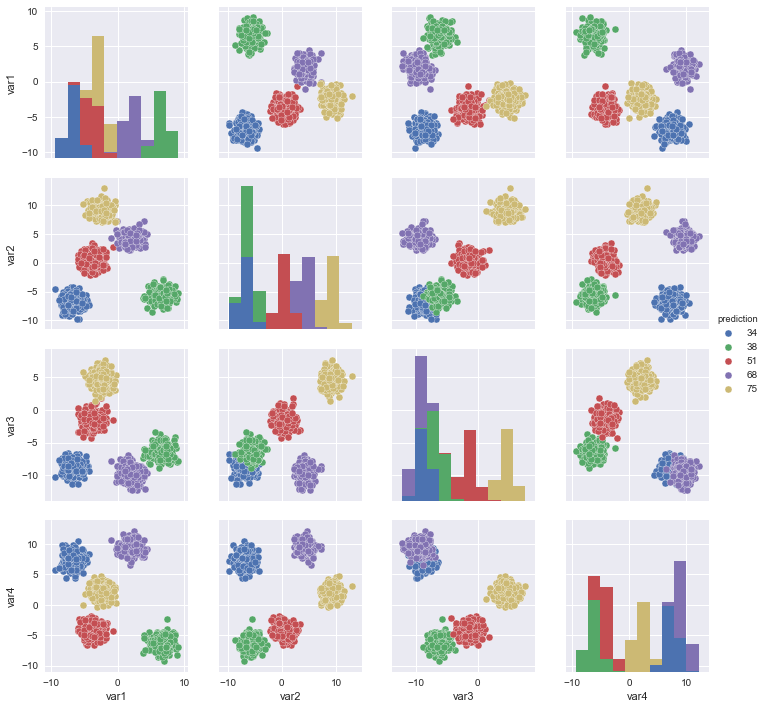
install.packages("CRPClustering") library(CRPClustering) result <- crp_gibbs(as.matrix(X), mu=c(0,0,0,0), sigma_table=14, alpha =0.3, ro_0 =0.1, burn_in=40, iteration =200) crp_graph_2d(as.matrix(X), result)
- 出力
[1] "Center coordinates of Clusters" cluster mu_k.1 mu_k.2 mu_k.3 mu_k.4 1 33 -1.7320941 -3.9238190 -0.02343495 -0.64889908 2 8 -5.4245621 -5.9558809 -2.48482642 4.09809942 3 5 -5.7573034 -1.9062441 -11.68433652 -0.60945428 4 44 1.8258171 -0.3260943 -0.48906236 1.01262547 5 9 -0.5258223 -11.0697677 -13.77974419 2.86106750 6 30 -4.2536066 -3.3689614 3.33196781 -2.17071835 7 29 -7.5764670 -3.0255271 -4.04623234 6.15198519 8 23 3.9127942 -1.5834261 -4.18575549 0.08683102 9 34 -2.9826239 -3.4543564 1.57405714 -1.00104252 [1] "The Number of Total Clusters : 9" [1] "Entropy All : 2.40723270175617" [1] "z_result has numbers of clusters."
library(MASS) dat <- rbind(mvrnorm(50, c(0,0), diag(c(1,1))), mvrnorm(50, c(2,2), diag(c(1,1))), mvrnorm(50, c(7,7), diag(c(1,1))), mvrnorm(50, c(8,9), diag(c(1,1)))) plot(dat,col=c(rep("red",50),rep("blue",50),rep("yellow",50),rep("black",50))) result <- crp_gibbs(as.matrix(dat), mu=c(0,0), sigma_table=14, alpha =0.3, ro_0 =0.1, burn_in=40, iteration =200)
library(mclust) Mclust(X) -> out plot(out)
- 今日のまとめとして高次元データ用のクラスタリング法のレビューを読んでみる(こちら)
- parsimonious gaussian mixture model をRのpgmm パッケージでやってみる
- 階層的クラスタリングをしつつ、どこで切るのがよいか教えてくれる
> str(gaelle.bclust) List of 14 $ merge : num [1:54, 1:2] -25 1 -12 -21 -8 5 2 4 -32 9 ... $ height : num [1:54] 0 14.3 26 37.8 49.6 ... $ order : num [1:55] 2 3 45 46 44 42 41 40 47 39 ... $ logposterior : num [1:54] -2931 -2916 -2905 -2893 -2881 ... $ clust.number : int [1:54] 54 53 52 51 50 49 48 47 46 45 ... $ cut : num 741 $ optim.alloc : int [1:55] 1 1 1 2 2 2 2 2 2 2 ... $ optim.clustno : int 7 $ data : num [1:55, 1:43] 0.33321 -0.00103 -0.2521 -0.17771 -0.43585 ... ..- attr(*, "dimnames")=List of 2 .. ..$ : chr [1:55] "ColWT.1" "ColWT.3" "ColWT.2" "d172.1" ... .. ..$ : chr [1:43] "maltose.MX1" "L.ascorbic" "glutamic.3" "raffinose2" ... $ repno : int [1:55] 1 1 1 1 1 1 1 1 1 1 ... $ transformed.par: num [1:6] -1.84 -0.99 1.63 0.08 -0.16 -1.68 $ labels : chr [1:55] "ColWT.1" "ColWT.3" "ColWT.2" "d172.1" ... $ effect.family : chr "gaussian" $ var.select : logi TRUE - attr(*, "class")= chr [1:2] "bclustvs" "hclust"